R code demo of
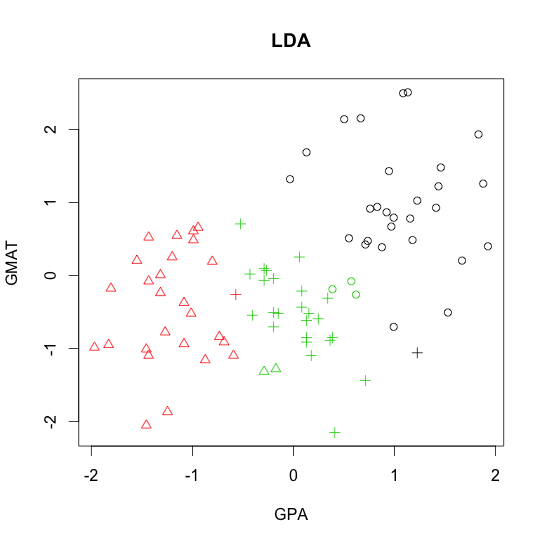
- Linear discriminant analysis (LDA)
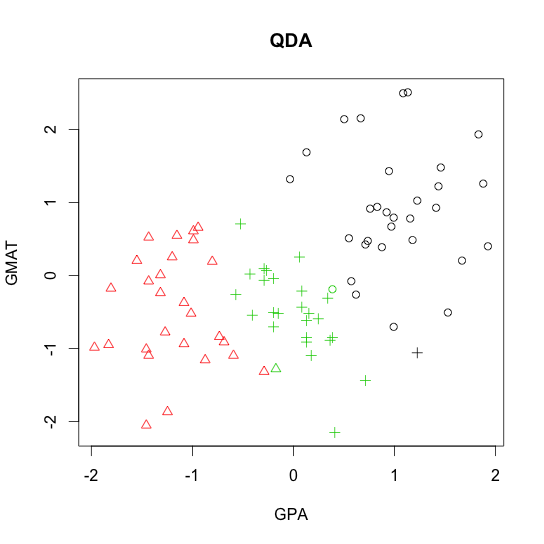
- Quadratic discriminant analysis (QDA)
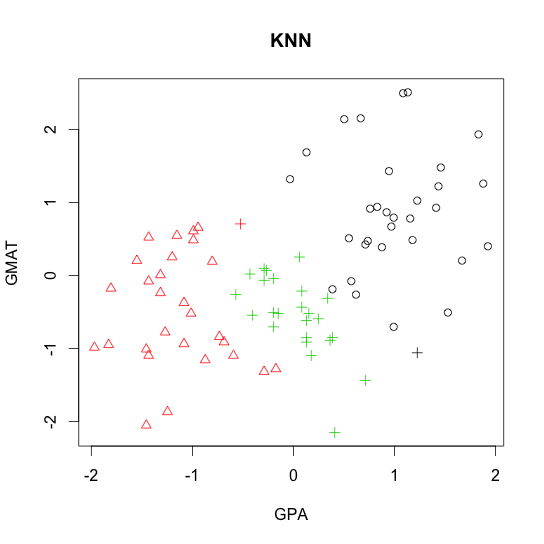
- k-nearest neighbor (KNN)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | |
1 2 3 4 5 6 7 8 | |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | |
R code demo of
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | |

1 2 3 4 5 6 7 8 | |

1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | |
