Comparative Studies of de novo Assembly Tools for Next-generation Sequencing Technologies
Yong Lin, Jian Li, Hui Shen, Lei Zhang, Christopher J Papasian and Hong-Wen Deng
Motivation: Several new de novo assembly tools have been developed recently to assemble short sequencing reads generated by next-generation sequencing platforms. However, the performance of these tools under various conditions has not been fully investigated, and sufficient information is not currently available for informed decisions to be made regarding the tool that would be most likely to produce the best performance under a specific set of conditions.
Results: We studied and compared the performance of commonly used de novo assembly tools specifically designed for next-generation sequencing data, including SSAKE, VCAKE, Euler-sr, Edena, Velvet, ABySS and SOAPdenovo. Tools were compared using several performance criteria, including N50 length, sequence cover-age, and assembly accuracy. Various properties of read data, including single-end/paired-end, sequence GC content, depth of coverage and base calling error rates, were investigated for their effects on the performance of different assembly tools. We also compared the computation time and memory usage of these seven tools. Based on the results of our comparison, the relative perform-ance of individual tools are summarized and tentative guidelines for optimal selection of different assembly tools, under different condi-tions, are provided.
拿流行的几个de novo assembly tools for next-generation sequencing做了系统的比较,是一篇很不错的文章。曾经有多少人徘徊于各种assembly软件中,不知道选择哪一种,这篇文章提供了很好的帮助。
作者比较了SSAKE, VCAKE, Euler-sr, Edena, Velvet, ABySS and SOAPdenovo,(竟然没有ALLPATHS-LG!)。
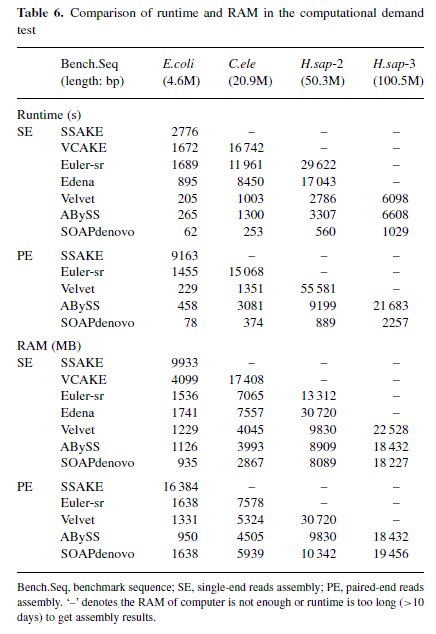
先用大到人类(其实最多只用了100Mb序列片段)小到E. coli的不同GC含量的基因组,模拟出single-end和paired-end、以及不同测序深度、不同测序片段长度的短序列片段,然后使用各个软件拼接,缺省设置,在各有2.40GB双核+12GB的8个nodes上运行,然后比较拼接结果:N50、基因组覆盖率(sequence coverage)、拼接错误(assembly error)和base calling error rate (BCER)等,还有软件运行时的硬件消耗。
最终的结论:(结论比较复杂,不同条件和要求下,各个软件表现有些差异。总的来看ABySS和SOAPdenovo比较“万金油”。)
再看非常重要的硬件消耗(还是SOAPdenovo和abyss给力啊,看来我眼光不错,我就是用的这两个,还有ALLPATHS-LG!)
本人看法:SOAPdenovo能拼接scaffolds,能产生更长的N50,而ABySS只能拼接到contigs;但ABySS基于MPI并行,对硬件需求低。ALLPATHS-LG和SOAPdenovo相似。